Google Deepmind has taken the wraps off a new version AlphaFold, their transformative machine learning model that predicts the shape and behavior of proteins. AlphaFold 3 is not only more accurate, but predicts interactions with other biomolecules, making it a far more versatile research tool — and the company is putting a limited version of the model free to use online.
From the debut of the first AlphaFold back in 2018, the model has remained the leading method of predicting protein structure from the sequence of amino acids that make them up.
Though this sounds like rather a narrow task, it’s foundational to nearly all biology to understand proteins — which perform a nearly endless variety of tasks in our bodies — at the molecular level. In recent years, computational modeling techniques like AlphaFold and RoseTTaFold have taken over from expensive, lab-based methods, accelerating the work of thousands of researchers across as many fields.
But the technology is still very much a work in progress, with each model “just a step along the way,” as Deepmind founder Demis Hassabis put it in a press call about the new system. The company teased the release late last year but this marks its official debut.
I’ll let the science blogs get into exactly how the new model improves outcomes, but suffice it here to say that a variety of improvements and modeling techniques have made AlphaFold 3 not just more accurate, but more widely applicable.
One of the limitations of protein modeling is that even if you know how the shape a sequence of amino acids will take, that doesn’t mean you necessarily know what other molecules it will bind to, and how. And if you want to actually do things with these molecules, which most do, you needed to find that out through more laborious modeling and testing.
“Biology is a dynamic system, and you have to understand how properties of biology emerged through the interactions between different molecules in the cell. And you can think of AlphaFold 3 as our first big step towards that,” Hassabis said. “It’s able to model proteins interacting, of course, with other proteins, but also other biomolecules, including, importantly DNA and RNA strands.”
AlphaFold 3 allows multiple molecules to be simulated at once — for example, a strand of DNA, some DNA-binding molecules, and perhaps some ions to spice things up. Here’s what you get for one such specific combination, with the DNA ribbons going up the middle, the proteins glomming onto the side, and I think those are the ions nestled in the middle there like little eggs:
This, of course, isn’t a scientific discovery in and of itself. But even to figure out that an experimental protein would bind at all, or in this way, or contort to this shape, was generally the work of days at the least or perhaps weeks to months.
While it’s difficult to overstate the excitement in this field over the last few years, researchers have largely been hamstrung by the lack of interaction modeling (of which the new version offers a form) and difficulty deploying the model.
This second issue is perhaps the greater of the two, as while the new modeling techniques were “open” in some sense, like other AI models they are not necessarily simple to deploy and operate. That’s why Google Deepmind is offering AlphaFold Server, a free, fully hosted web application making the model available for non-commercial use.
It’s free and quite easy to use — I did it in another window on the call while they were explaining it (which is how I got the image above). You just need a Google account, and then you feed it as many sequences and categories as it can handle — there are some examples provided — and submit; in a few minutes your job should be done and you’ll be given a live 3D molecule colored to represent the model’s confidence in the conformation at that position. As you can see in the one above, the tips of the ribbons and those parts more exposed to rogue atoms are lighter or red to indicate less confidence.
I asked whether there was any real difference between the publicly available model and the one being used internally; Hassabis said that “We’ve made the majority of the new model’s capabilities available,” but didn’t elaborate beyond that.
It’s clearly Google throwing its weight about — while to a certain extent, keeping the best bits for themselves, which of course is their prerogative. Making a free, hosted tool like this involves dedicating considerable resources to the task — make no mistake, this is a money pit, an expensive (to Google) shareware version to convince the researchers of the world that AlphaFold 3 should be, at the very least, an arrow in their quiver.
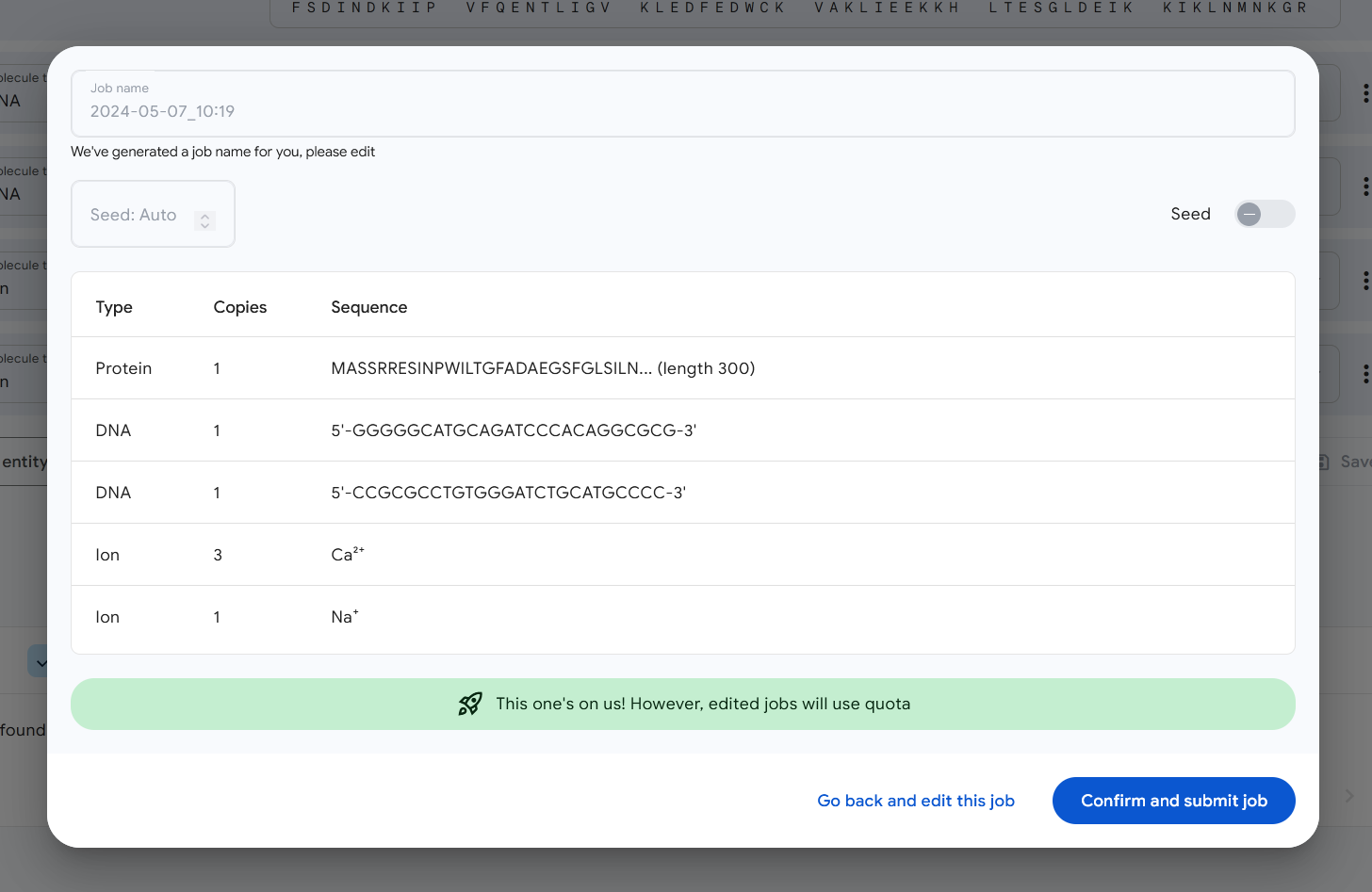
Image Credits: Google Deepmind
That’s all right, though, because the tech will likely print money through Alphabet subsidiary (which makes it Google’s… cousin?) Isomorphic Labs, which is putting computational tools like AlphaFold to work in drug design. Well, everyone is using computational tools these days — but Isomorphic got first crack at Deepmind’s latest models, combining it with “some more proprietary things to do with drug discovery,” as Hassabis noted. The company already has partnerships with Eli Lilly and Novartis.
AlphaFold isn’t the be-all and end-all of biology, though — just a very useful tool, as countless researchers will agree. And it allows them to do what Isomorphic’s Max Jaderberg called “rational drug design.”
“If we think about, day to day, how this has an impact at Isomorphic labs: it allows our scientists, our drug designers, to create and test hypotheses at the atomic level, and then within seconds produce highly accurate structure predictions… to help the scientists reason about what are the interactions to make, and how to advance those designs to create a good drug,” he said. “This is compared to the months or even years it might take to do this experimentally.”
While many will celebrate the accomplishment and the wide availability of a free, hosted tool like AlphaFold Server, others may rightly point out that this isn’t really a win for open science.
Like many proprietary AI models, AlphaFold’s training process and other information crucial to replicating it — a fundamental part of the scientific method, you will recall — are largely and increasingly withheld. While the paper published in Nature does go over the methods of its creation in some detail, a lot of important details and data are lacking, meaning scientists who want to use the most powerful molecular biology tool on the planet will have to do so under the watchful eye of Alphabet, Google, and Deepmind (who knows which actually holds the reins).
Open science advocates have said for years that, while these advances are remarkable, it is always better in the long run to share this kind of thing openly. That is, after all, how science moves forward, and indeed how some of the most important software in the world has evolved as well.
Making AlphaFold Server free to any academic or non-commercial application is in many ways a very generous act. But Google’s generosity seldom comes no strings attached. No doubt many researchers will nevertheless take advantage of this honeymoon period to use the model as much as humanly possible before the other shoe drops.




